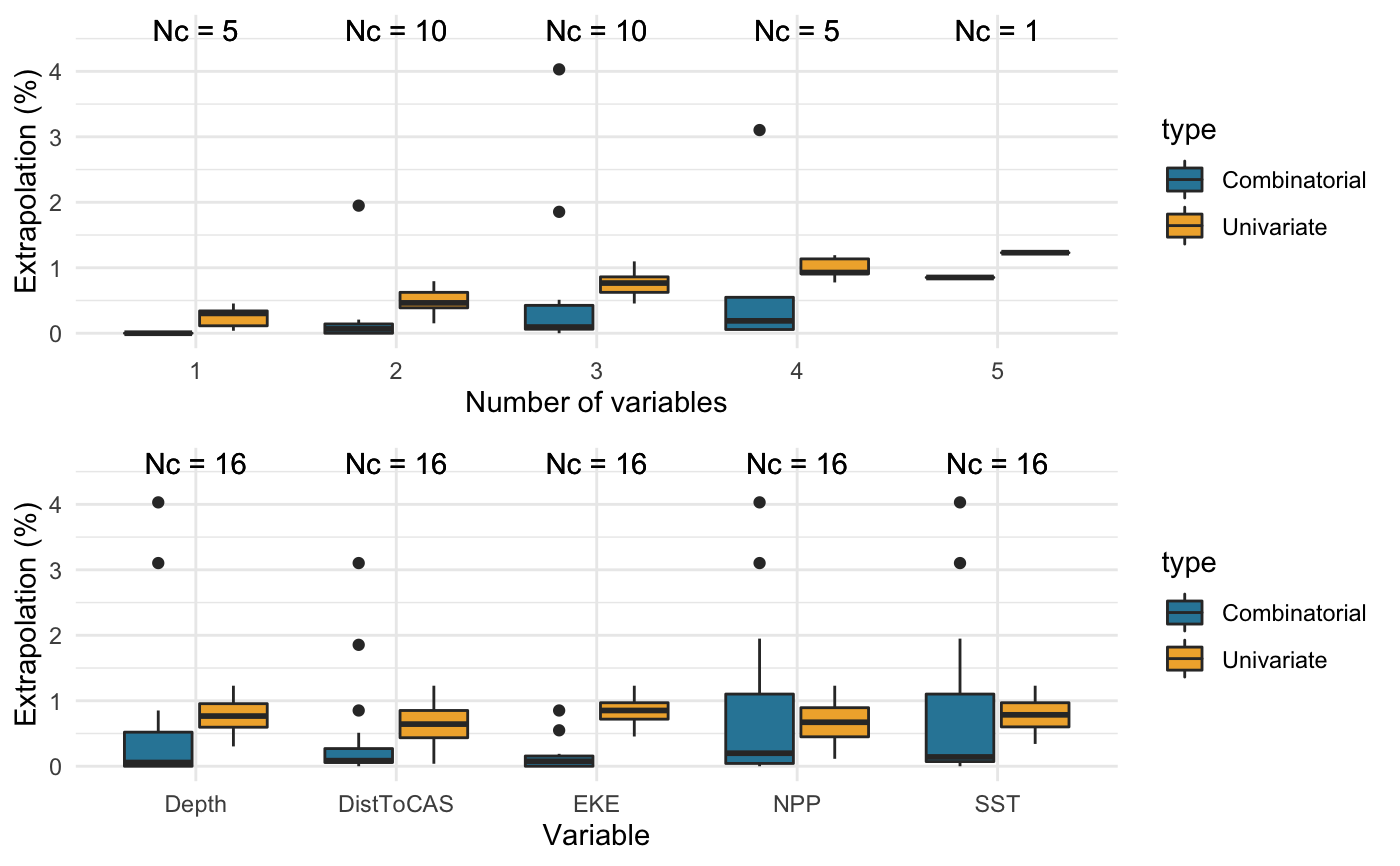
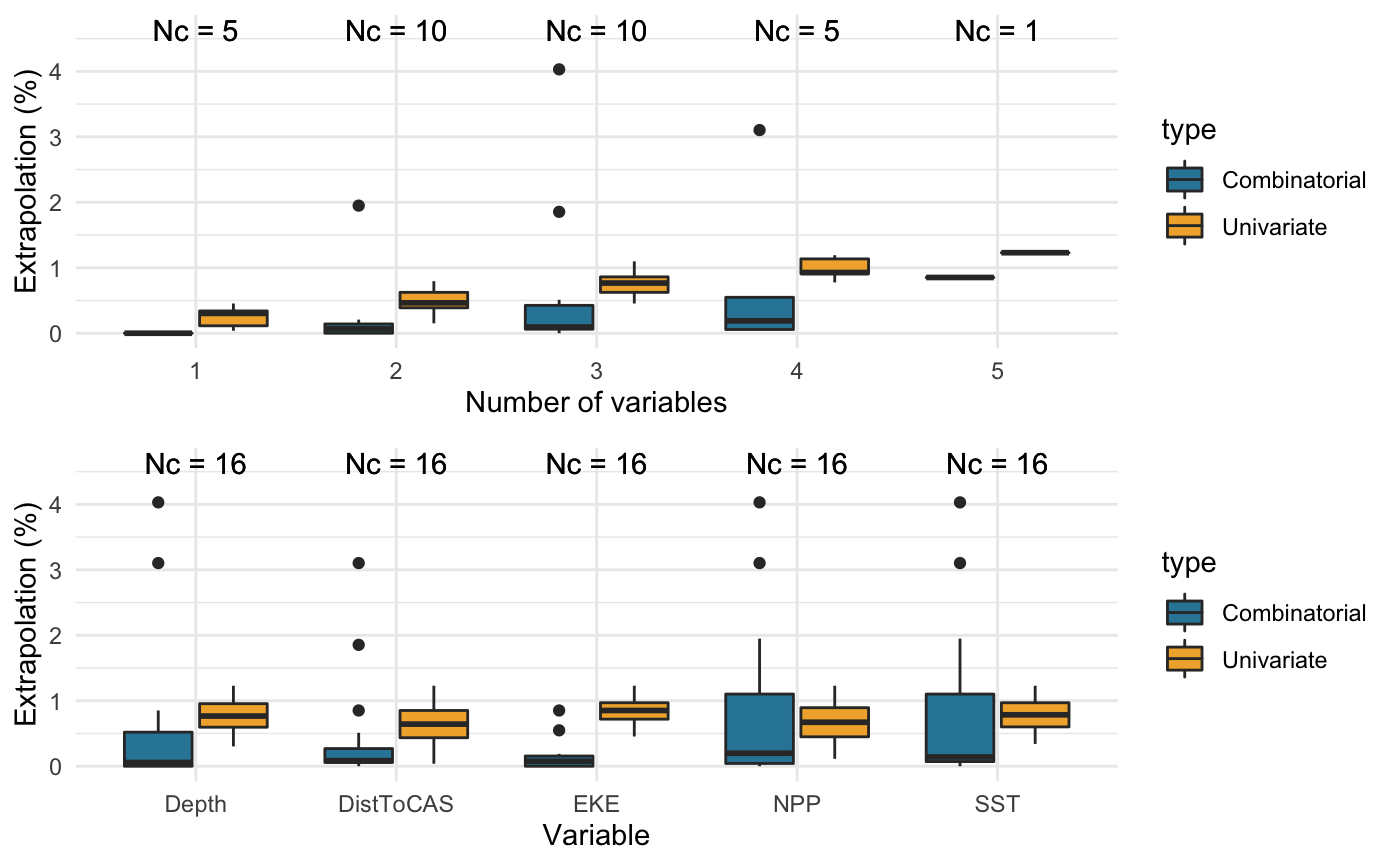
Compare different combinations of covariates
compare_covariates.RdSummarises the extent of univariate (Type I) and combinatorial (Type II) extrapolation associated with different combinations of input covariates.
compare_covariates( extrapolation.type = "both", extrapolation.object = NULL, n.covariates = NULL, create.plots = TRUE, display.percent = TRUE, verbose = TRUE, ... )
Arguments
| extrapolation.type | Character string. Type of extrapolation to be assessed. Can be one of |
|---|---|
| extrapolation.object | List object as returned by compute_extrapolation. |
| n.covariates | Maximum number of covariates. The function will compare all combinations of 1 to |
| create.plots | Logical, defaults to |
| display.percent | Logical. If |
| verbose | Logical. Show or hide possible warnings and messages. |
| ... | Additional parameters passed to compute_extrapolation. These are optional when |
Value
Prints a summary table in the R console. Also generates summary boxplots if create.plots is set to TRUE.
Details
The extent and magnitude of extrapolation naturally vary with the type and number of covariates considered. It may be useful, therefore, to test different combinations of covariates to inform their selection a priori, i.e. before model fitting, thereby supporting model parsimony.
References
Bouchet PJ, Miller DL, Roberts JJ, Mannocci L, Harris CM and Thomas L (2019). From here and now to there and then: Practical recommendations for extrapolating cetacean density surface models to novel conditions. CREEM Technical Report 2019-01, 59 p. https://research-repository.st-andrews.ac.uk/handle/10023/18509
Mesgaran MB, Cousens RD, Webber BL (2014). Here be dragons: a tool for quantifying novelty due to covariate range and correlation change when projecting species distribution models. Diversity & Distributions, 20: 1147-1159. DOI: 10.1111/ddi.12209
See also
Examples
library(dsmextra) # Load the Mid-Atlantic sperm whale data (see ?spermwhales) data(spermwhales) # Extract the data segs <- spermwhales$segs predgrid <- spermwhales$predgrid # Define relevant coordinate system my_crs <- sp::CRS("+proj=aea +lat_1=38 +lat_2=30 +lat_0=34 +lon_0=-73 +x_0=0 +y_0=0 +datum=WGS84 +units=m +no_defs +ellps=WGS84 +towgs84=0,0,0") # Define covariates my_cov <- c("Depth", "SST", "NPP", "DistToCAS", "EKE") # Compare the extent of univariate and combinatorial # extrapolation for all combinations of 1 to 5 covariates compare_covariates(extrapolation.type = "both", covariate.names = my_cov, n.covariates = NULL, samples = segs, prediction.grid = predgrid, coordinate.system = my_crs, create.plots = TRUE, display.percent = TRUE)#>#>#>#>#>#> #> #> Extrapolation Minimum n_min Maximum n_max #> -------------- ----------------- ------ -------------------------------- ------ #> Univariate DistToCAS 2 Depth, SST, NPP, DistToCAS, EKE 65 #> Combinatorial Depth 0 Depth, SST, NPP 213 #> SST 0 - - #> NPP 0 - - #> DistToCAS 0 - - #> EKE 0 - - #> Depth, DistToCAS 0 - - #> Depth, EKE 0 - - #> NPP, DistToCAS 0 - - #> NPP, EKE 0 - - #> Depth, SST, EKE 0 - - #> Depth, NPP, EKE 0 - - #> Both DistToCAS 2 Depth, SST, NPP 252# Can also run this function directly from the # object returned by compute_extrapolation spermw.extrapolation <- compute_extrapolation(samples = segs, covariate.names = my_cov, prediction.grid = predgrid, coordinate.system = my_crs)#>#>compare_covariates(extrapolation.type = "both", extrapolation.object = spermw.extrapolation)#>#>#>#>#>#> #> #> Extrapolation Minimum n_min Maximum n_max #> -------------- ----------------- ------ -------------------------------- ------ #> Univariate DistToCAS 2 Depth, SST, NPP, DistToCAS, EKE 65 #> Combinatorial Depth 0 Depth, SST, NPP 213 #> SST 0 - - #> NPP 0 - - #> DistToCAS 0 - - #> EKE 0 - - #> Depth, DistToCAS 0 - - #> Depth, EKE 0 - - #> NPP, DistToCAS 0 - - #> NPP, EKE 0 - - #> Depth, SST, EKE 0 - - #> Depth, NPP, EKE 0 - - #> Both DistToCAS 2 Depth, SST, NPP 252